| Search Protocol |
|---|
Search Summary
Quick Search: Users can make use of "Quick Search" function to query the interested information directly in "HOME" page (Figure 1).
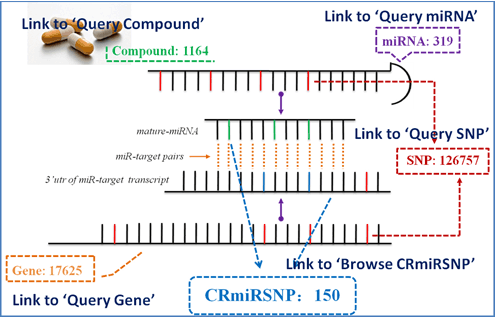
Figure 1. Quick search in HOME page.
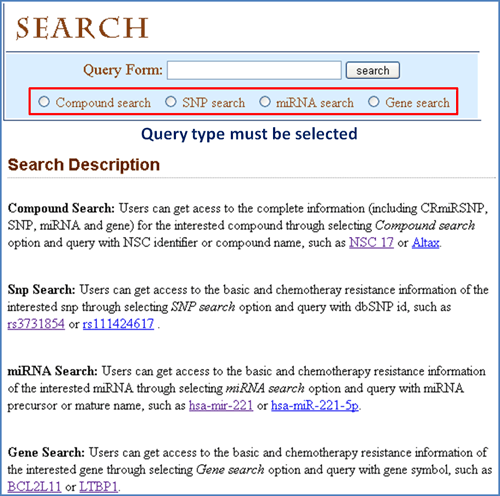
Figure 2. Complete search in SEARCH page.
Query Compound
Query Compound: Users can search the interested compound to access miRSNP, SNP, miRNA, and gene related to the chemotherapy resistance of the queried compound (Figure 3).
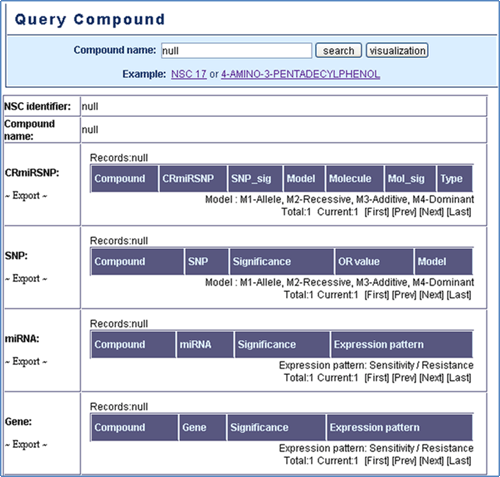
Figure 3. Query compound.
Query SNP
Query SNP: In the search result page, the users can know that whether the queried SNP is CRmiRSNP and which chemotherapy resistance of compound is associated with this SNP. Additionally, users can aware that whether the SNP presents in the 3'UTR of the gene transcripts or in miRNA precursor transcripts (Figure 4).
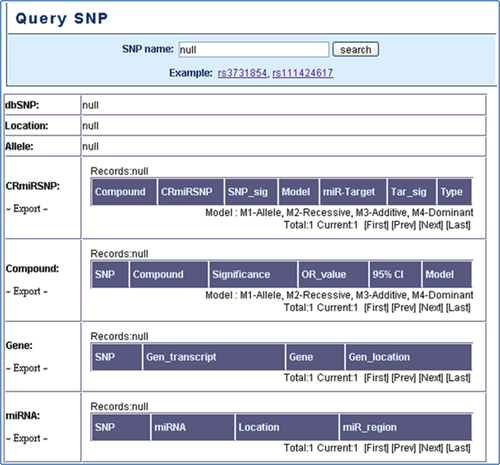
Figure 4. Query snp.
Query MiRNA
Query MiRNA: In the search result page, the users can know that whether the CRmiRSNPs locate on the queried miRNA and which compounds chemotherapy resistance is associated with this miRNA. Additionally, users can find the information about SNPs that present in this miRNA precursor transcript and target genes of this miRNA (Figure 5).
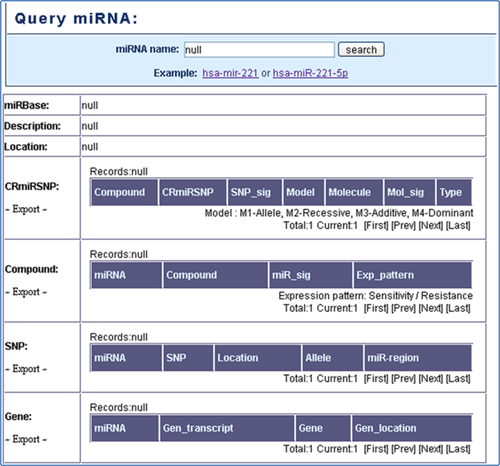
Figure 5. Query mirna.
Query Gene
Query Gene: In the search result page, the users can know that whether the CRmiRSNPs locate on 3'UTR of the queried gene transcripts and which compounds chemotherapy resistance is associated with this gene. Additionally, users can find the information about SNPs that present in 3'UTR of this gene's transcripts and miRNAs that potentially target this gene (Figure 6).

Figure 6. Query gene.
Genomic Distribution Map
Genomic Distribution Map: Clicking the 'visualization' button in the "Query Compound" page to access the Genomic Distribution Map for each compound (Figure 7).
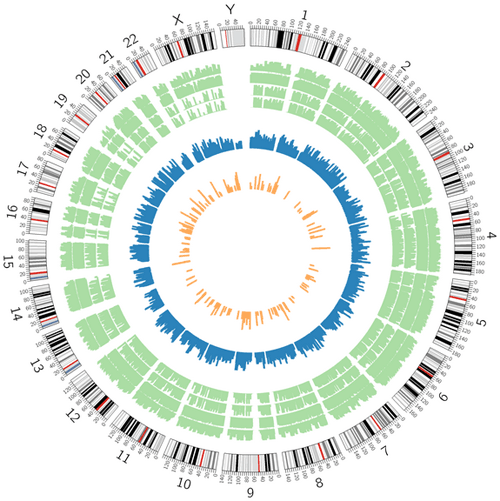
Figure 7. Genomic distribution map. Blue bars indicate chemotherapy resistance-associated genes, orange bars indicate chemotherapy resistance-associated miRNAs, and green bars represent chemotherapy resistance-associated SNPs by four models.
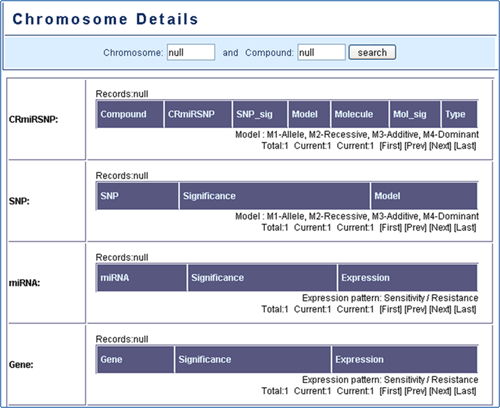
Figure 8. Chromosome details.