Manual
REMR (RNA Editing-Mediated miRNA Regulation) Database is an online platform designed to store and share RNA editing-mediated miRNA regulatory triplets across various cancer types. This database integrates the complex interactions between RNA editing and miRNA regulation, offering valuable insights for cancer research, prognosis evaluation, and drug response prediction.
The database is built upon the REMR algorithm, which utilizes a two-step filtering process to systematically identify how RNA editing mediates the regulatory relationship between miRNAs and their target genes:
- Step 1: Identifies RNA editing sites, particularly those occurring in miRNAs and 3'UTRs of target genes, that can alter miRNA binding sites.
- Step 2: Calculates conditional mutual information (cMI) to assess the dependence of miRNA-target gene regulatory relationships on RNA editing.
Using this process, the REMR database has identified numerous RNA editing-mediated miRNA regulatory triplets in ten major cancer types from the TCGA dataset.
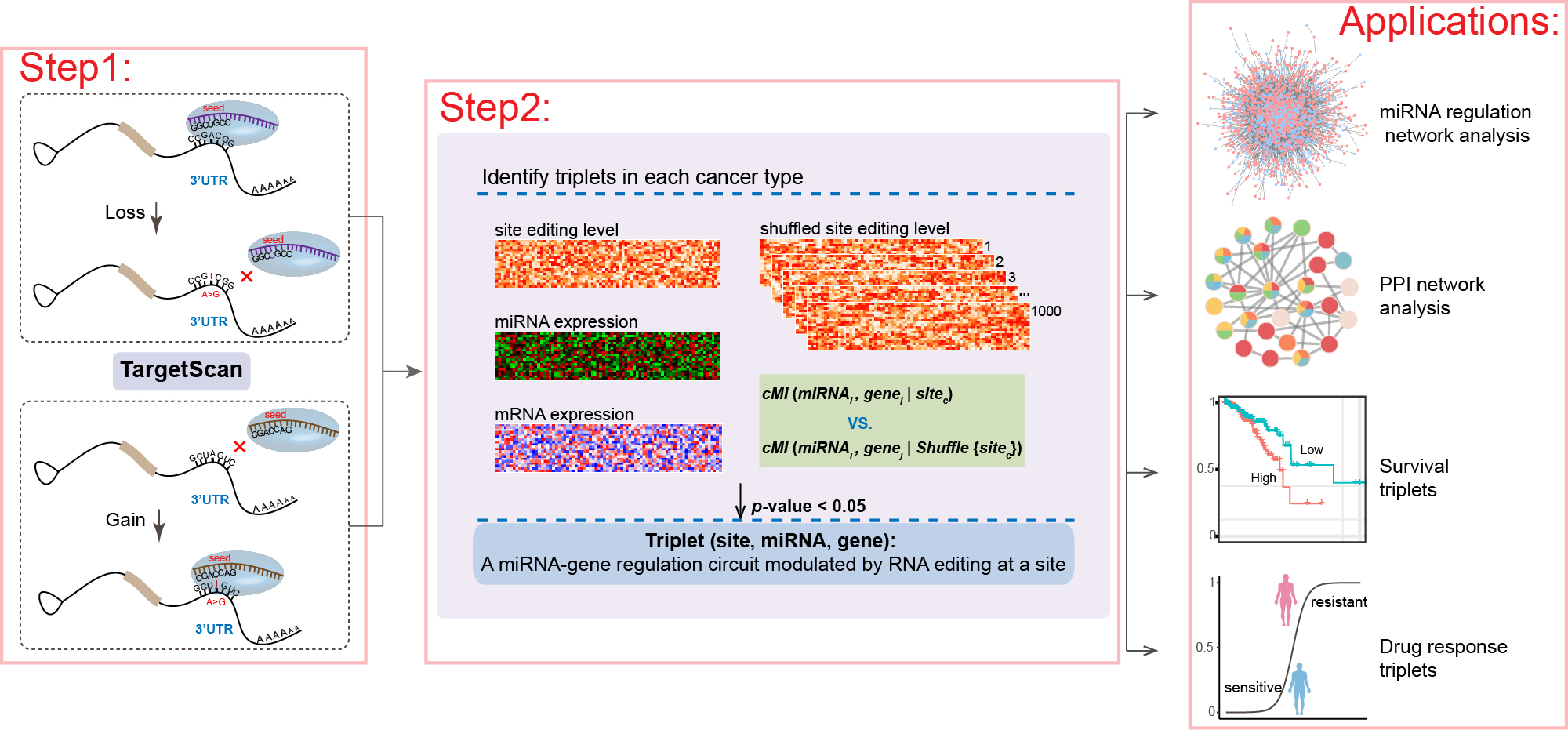
Workflow of REMR algorithm
The applications of REMR include:
- Systematically identifying and analyzing RNA editing-mediated miRNA regulatory networks in cancer.
- Exploring the underlying mechanisms of RNA editing in cancer progression and drug resistance.
- Identifying prognosis-related regulatory triplets.
- Identifying drug response-related regulatory triplets.
The REMR database utilizes the "Genome Browser" module (embedded with JBrowse) to visualize the genomic locations of RNA editing sites within triplets. In the Genome Browser interface, users can query triplet-related information in two ways:
(1).By entering a specific chromosomal position or region: Users can specify a genomic region to view RNA editing sites within that region and the triplet information associated with their regulatory roles.
- For example, first select a chromosome number, such as 5. Then, enter a genomic interval, such as the start site "79,923,318" and the end site "79,923,346" (Fig. 1), or directly input a single position, such as "79,923,332".
- Next, press the "Enter" button to search. You can right-click and open the feature detail (Fig. 2) to review detailed information about the RNA editing site-mediated miRNA regulation in cancer based on findings from our study (Fig. 3).
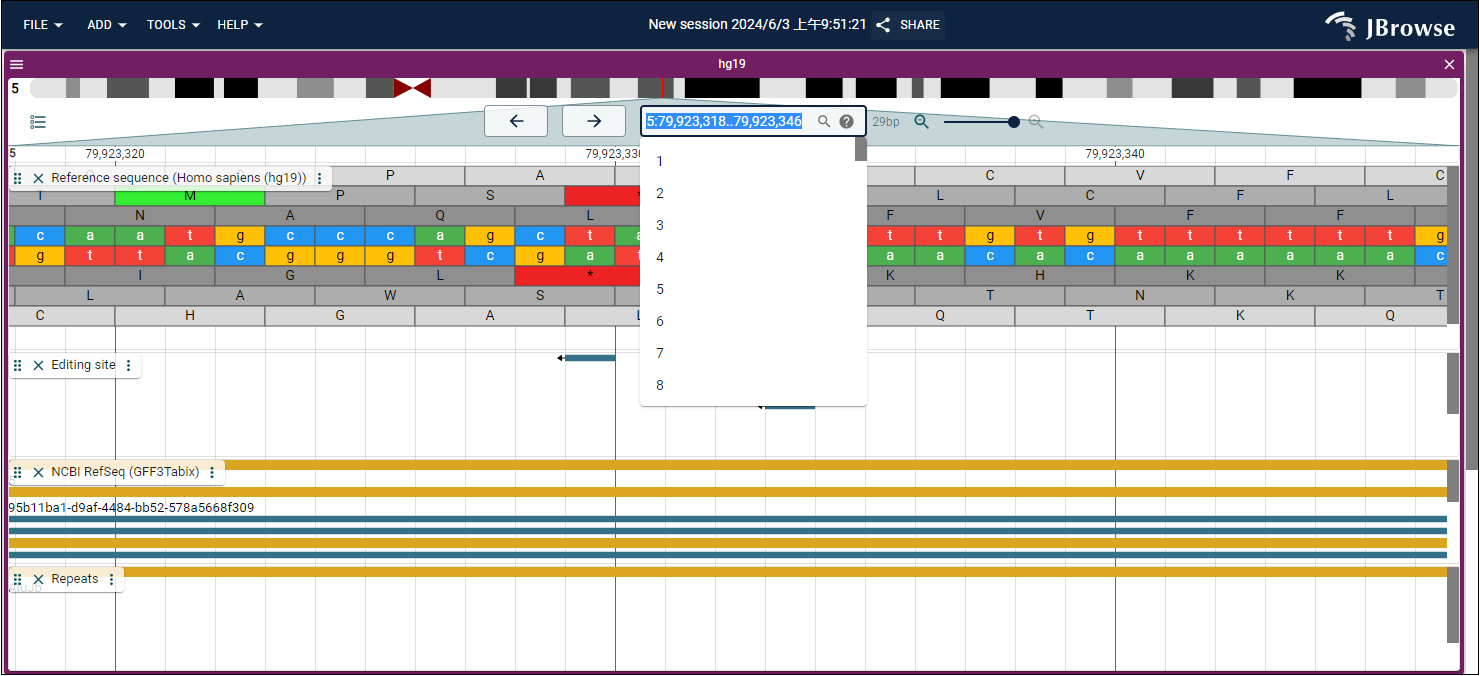
Fig.1 Search RNA editing site by genome locations in Gemome Browser
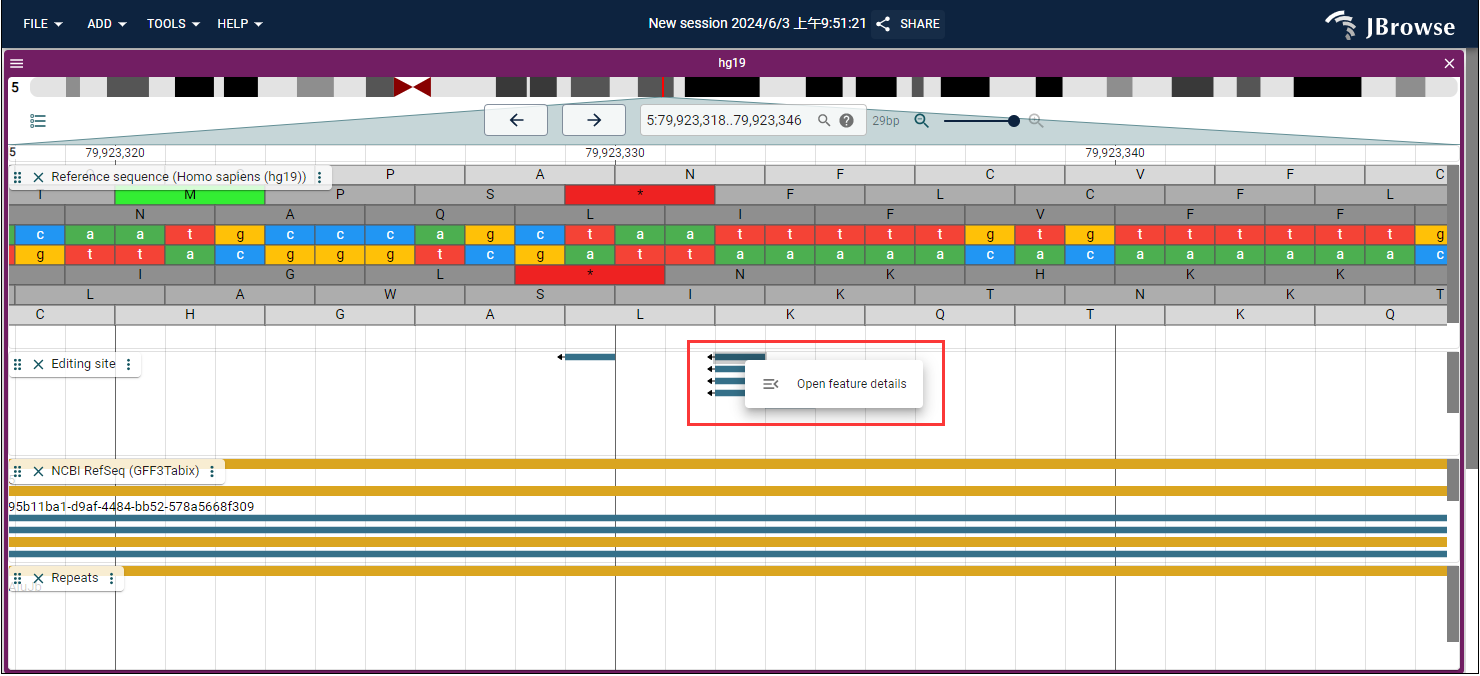
Fig.2 Open feature detail in Gemome Browser
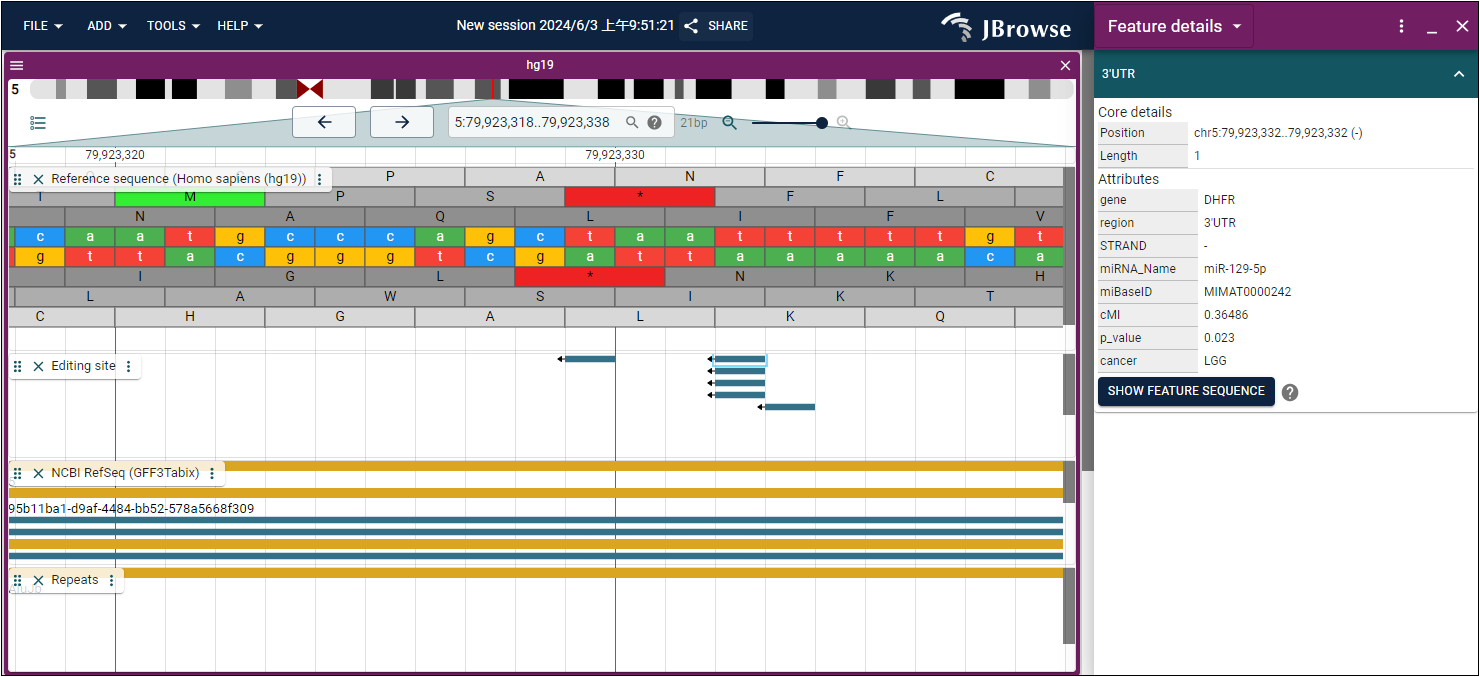
Fig.3 The miRNA regulation information of mediated by "chr5:79,923,332" in cancers.
(2).By entering a gene symbol: Users can input the symbol of a gene of interest to find RNA editing sites located on that gene and their involvement in triplet regulation.
- For example, you can directly enter a gene symbol into the search box to query information of interest, such as DHFR. Next, press the "Enter" button to search. You will then retrieve RNA editing sites on DHFR that affect miRNA regulation (Fig. 4)
- If you wish to view more detailed information on RNA editing-mediated miRNA regulation, you can follow similar steps as in Fig. 2 and Fig. 3 to access these details.
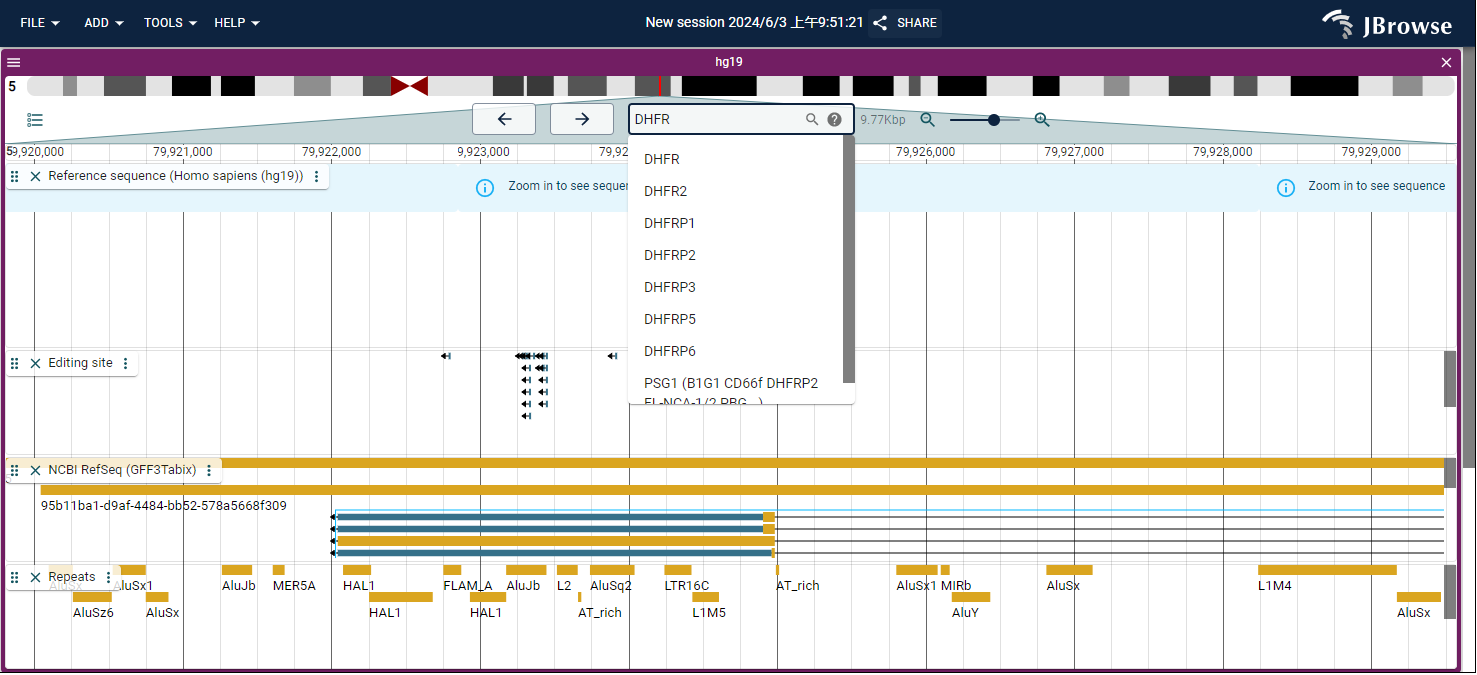
Fig.4 Search RNA editing sites by gene symbol in Gemome Browser
In particular, the reference genome used in our study is the hg19 version. The genome browser also provides transcriptome annotation information and genomic repeat region annotation information for the RNA editing sites. This additional data helps users gain a deeper understanding of the genomic context and functional significance of the RNA editing events.
In the REMR database, you can query all RNA editing-mediated miRNA regulatory triplets identified in this study. The "TRIPLETS" interface offers search options based on cancer type, RNA editing site, miRNA, and gene, allowing users to retrieve relevant information through either a single search criterion or a combination of multiple criteria. Below, we provide two examples to demonstrate the processes for both single and multi-criteria searches:
(1). single search criterion:
- Taking a single search for Bladder Urothelial Carcinoma (BLCA) as an example. In "TRIPLETS" interface, select "Bladder Urothelial Carcinoma (BLCA)" and then press the "Search" button (Fig. 5).The results will list all potential entries related to your inputted BLCA (Fig. 6).
- Furthermore, you can review more detailed information about the RNA editing site-mediated miRNA regulation by clicking on "more" (Fig. 7).
- In the page (Fig. 8), the TargetScan column shows how an edited site can result in the gain or loss of miRNA binding, along with the binding position details. The "seq.start" represents the starting position of the binding sites in a 101 bp sequence, which includes 50 bp upstream and 50 bp downstream of the editing site, while "seq.end" represents the ending position. Notably, the "+" strand sequence runs from left to right, while the "-" strand sequence runs from right to left. This orientation can be observed in the Genome Browser, where position "51" marks the RNA editing site.
Additionally, the column provides the type of miRNA binding predicted by TargetScan, the seed sequence information, and the sequence details of the mature miRNA. The embedded JBrowse tool is also used to visualize the genomic locations of RNA editing sites within the triplet. Further triplet details can be explored in the Genome Browser.
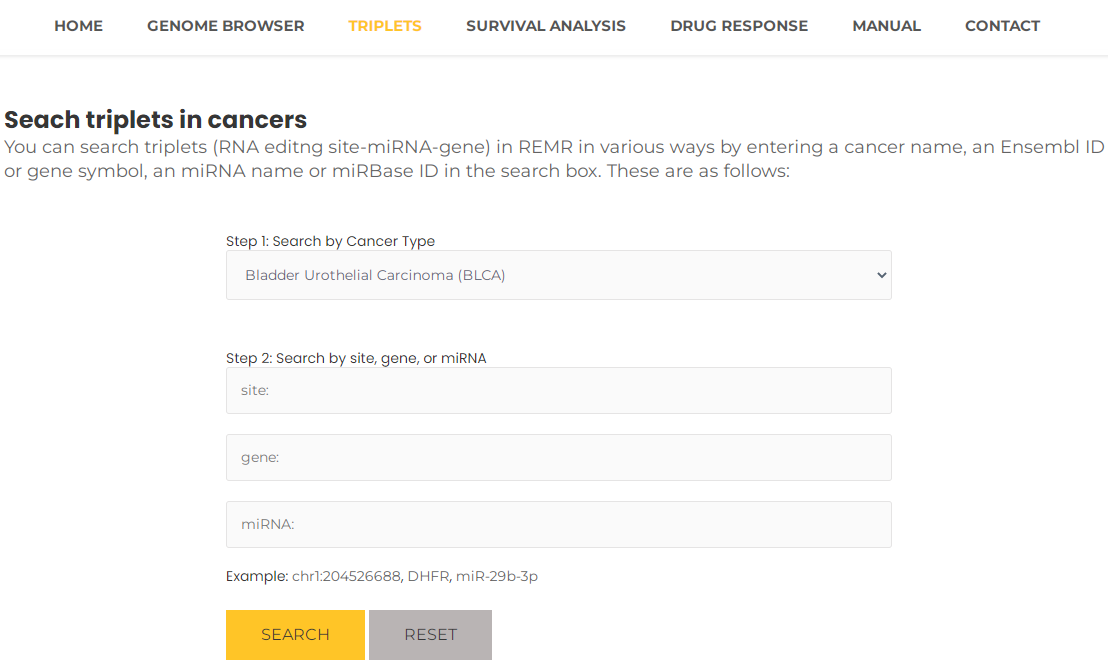
Fig.5 Search triplets by cancer name.
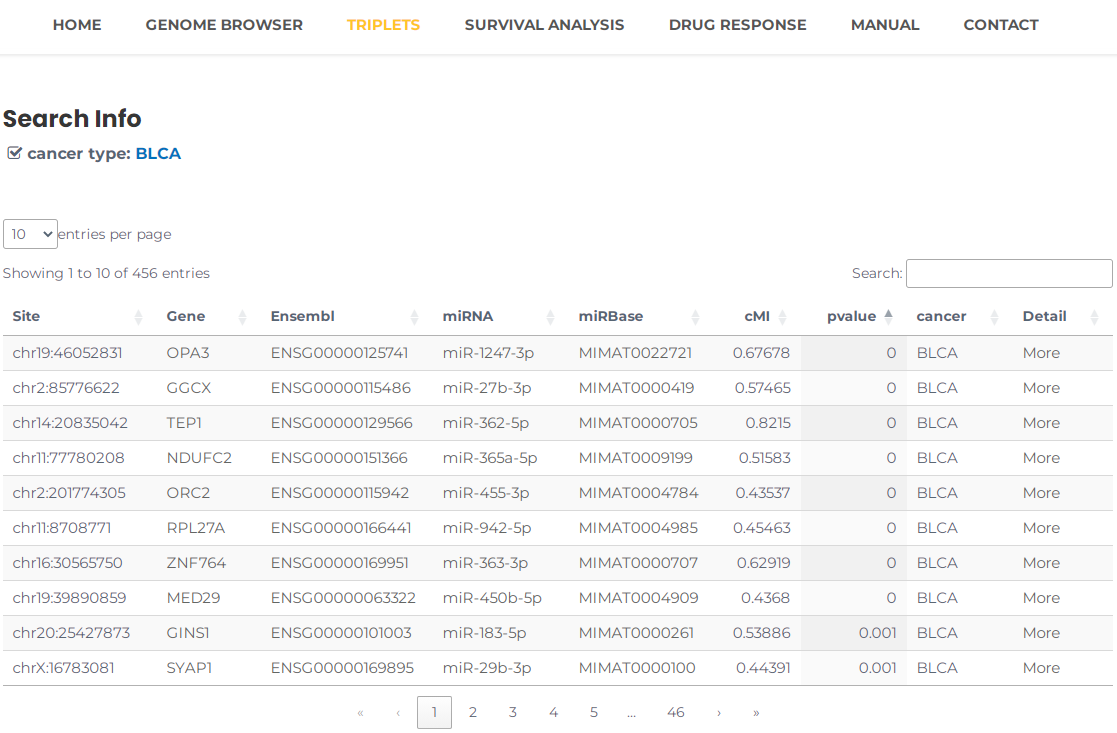
Fig.6 The result of "BLCA".
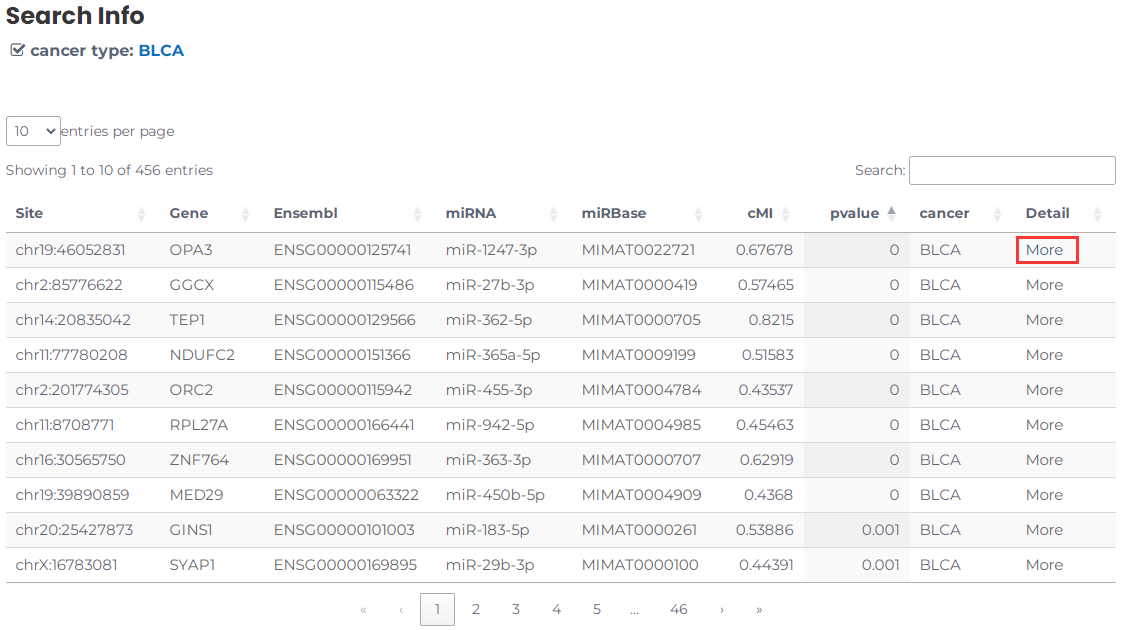
Fig.7 Click on the more.
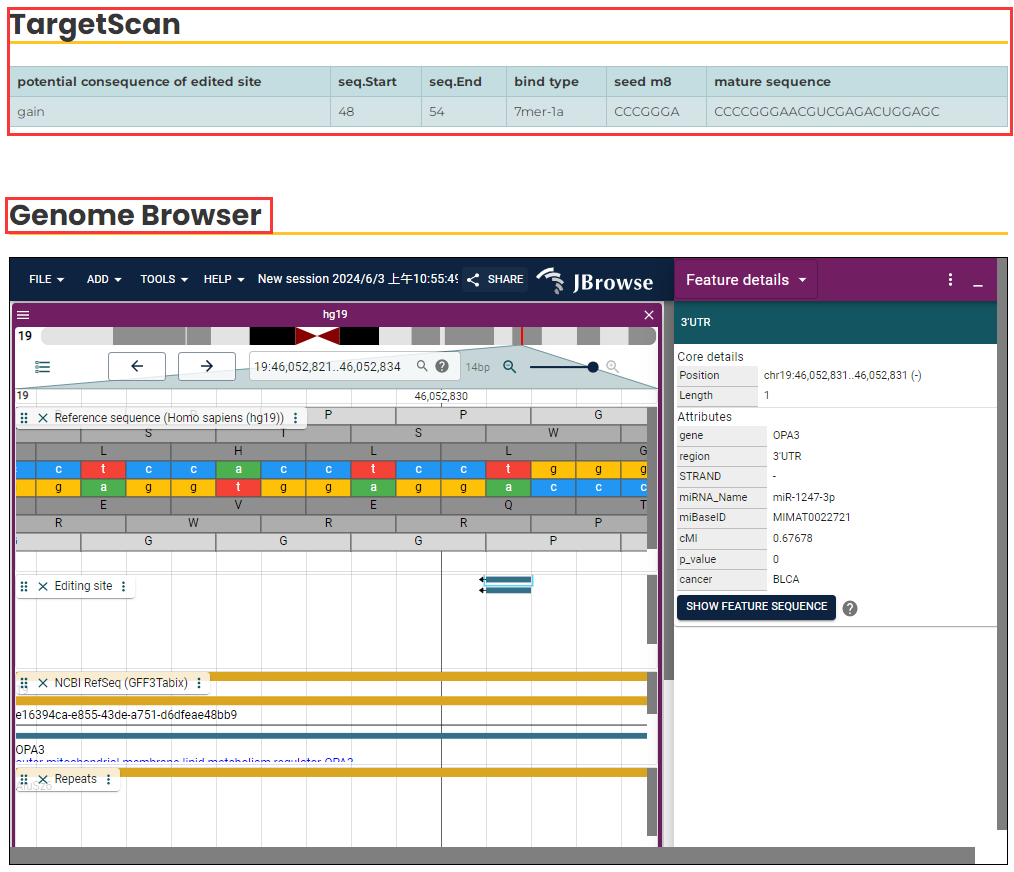
Fig.8 Detailed information of triplet.
(2). combination of multiple criteria:
- If you want to query a RNA editing site that mediates miRNA regulation in a particular cancer, for example, chr1:204526688 and BLCA, you can select BLCA and input chr1:204526688, then press the "Search" button (Fig. 9). The results will list all potential entries related to your input (Fig. 10). Similar to the description above , by clicking on the "more" button, you can obtain more detailed information about the triplet.
- Similarly, you can query a gene to review the triplets associated with it in a particular cancer, for example, DHFR (or Ensembl ID, ENSG00000228716) and BLCA, you can select BLCA and input DHFR, then press the "Search" button (Fig. 11). The results will list all potential entries related to your input (Fig. 12).
- Similarly, you can query a miRNA to review the triplets associated with it in a particular cancer, for example, miR-29b-3p (or miRBase ID, MIMAT0000100) and BLCA, you can select BLCA and input miR-29b-3p, then press the "Search" button (Fig. 13). The results will list all potential entries related to your input (Fig. 14).
- You can query matching results by entering combinations of site, miRNA, gene, and cancer type according to your needs. For example, you can select DHFR and miR-129-5p, then press the "Search" button (Fig. 15). The results will list all potential entries related to your input (Fig. 16).
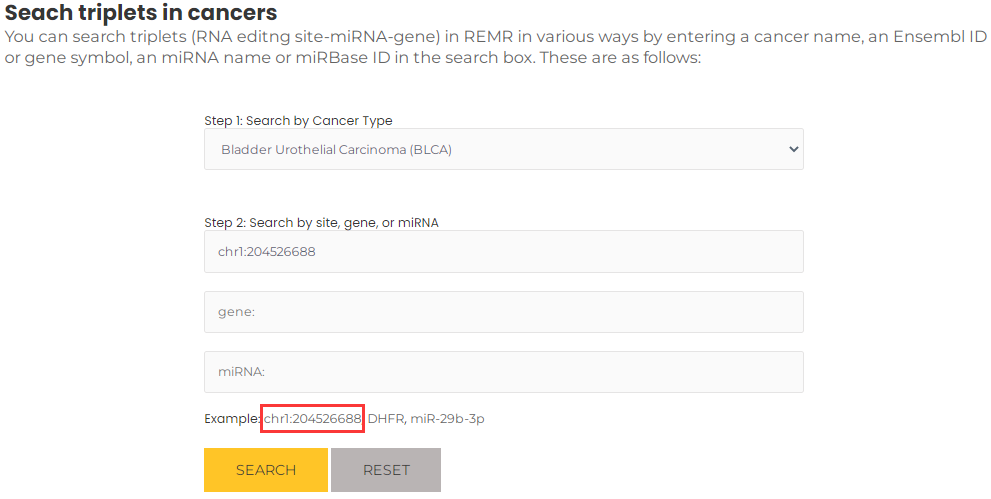
Fig.9 Search triplets by a RNA editing site and a particular cancer.
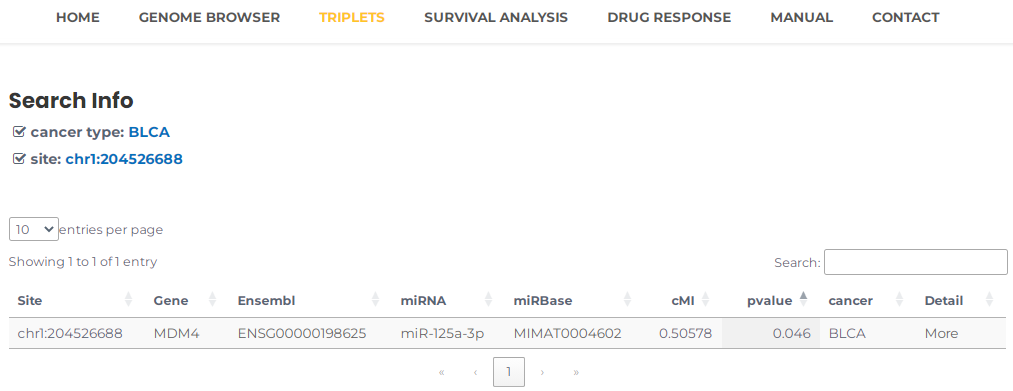
Fig.10 The result of "chr1:204526688" and "BLCA".
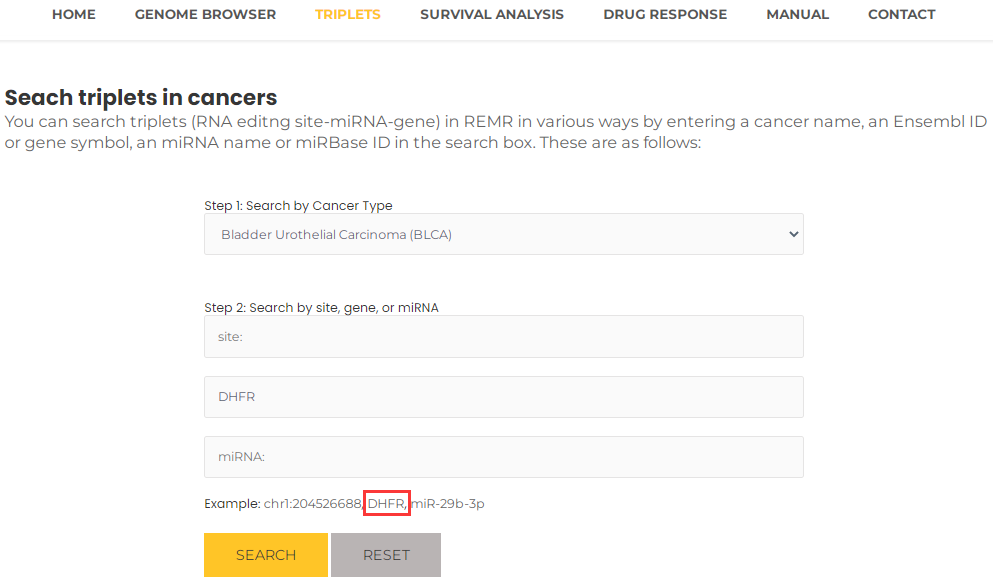
Fig.11 Search triplets by a gene and a particular cancer.
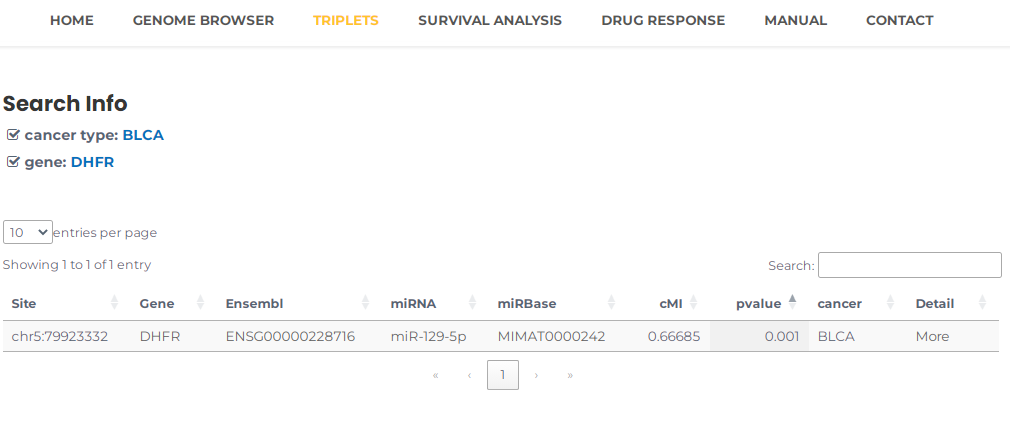
Fig.12 The result of "DHFR" and "BLCA".
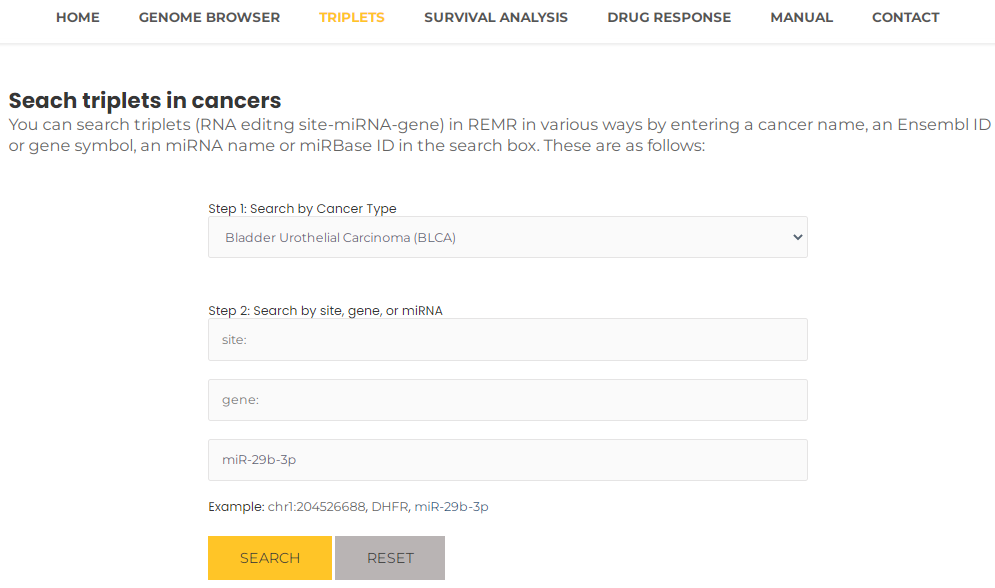
Fig.13 Search triplets by a miRNA and a particular cancer.
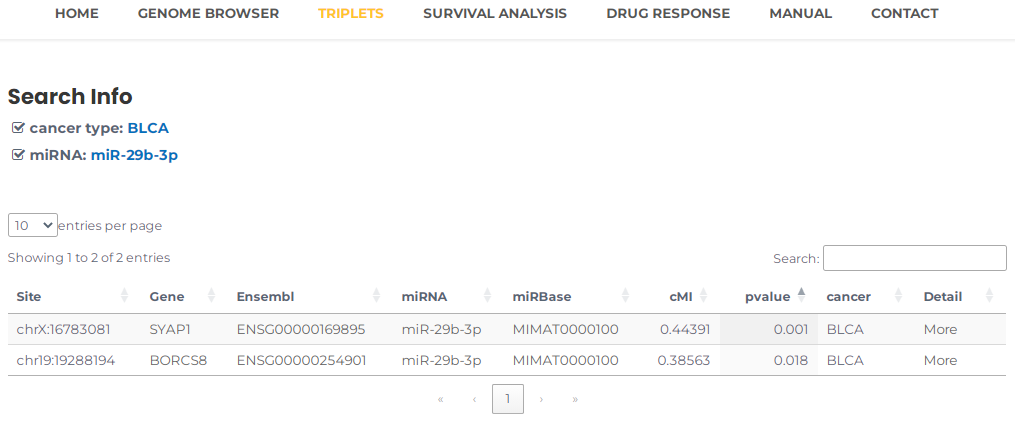
Fig.14 The result of "miR-29b-3p" and "BLCA".
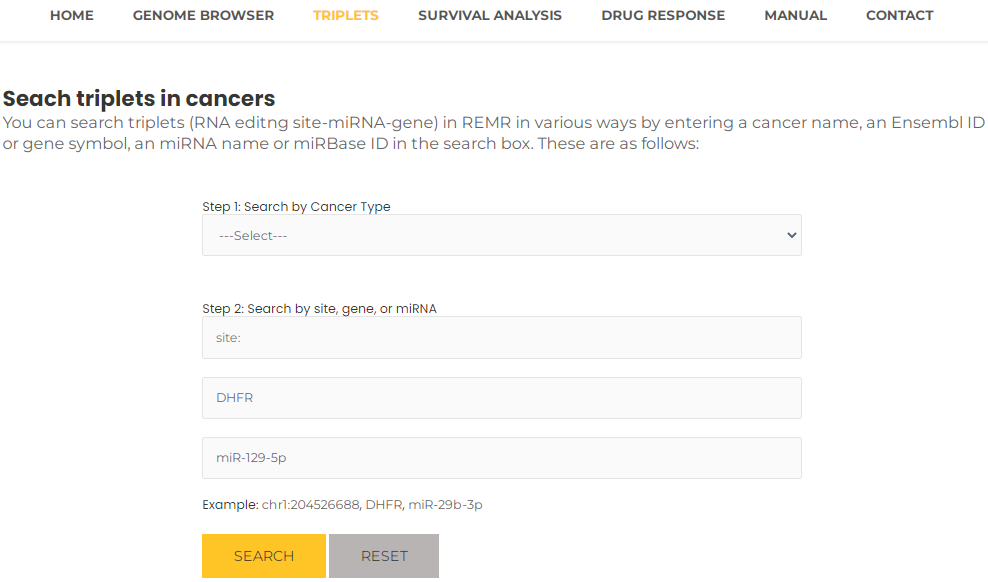
Fig.15 Search triplets by gene and miRNA.
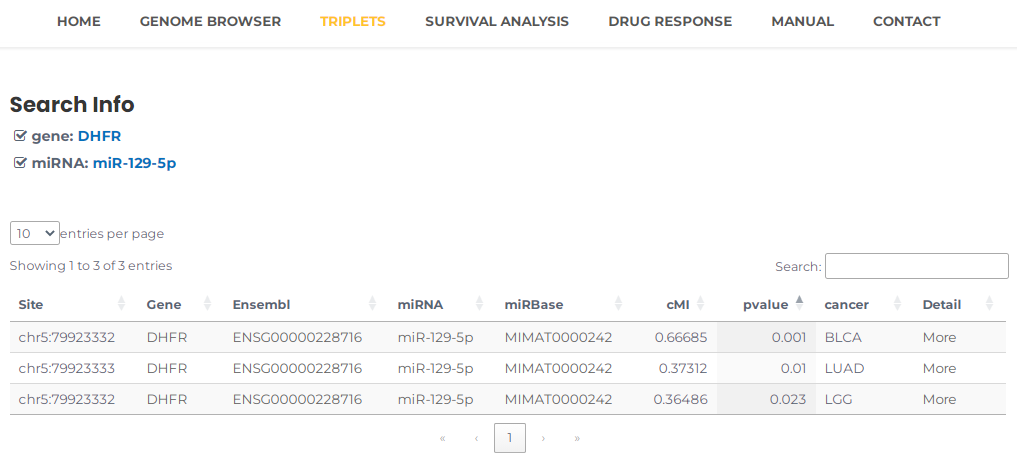
Fig.16 The result of "DHFR" and "miR-129-5p".
In REMR, you can review all identified prognosis-associated triplets in a particular cancer. The "SURVIVAL ANALYSIS" interface offers search options based on cancer type, RNA editing site, miRNA, and gene, allowing users to retrieve relevant information through either a single search criterion or a combination of multiple criteria.
- Taking a single search for Lower Grade Glioma (LGG) as an example. Select Lower Grade Glioma (LGG) and then press the "Search" button (Fig. 17). The results will list all potential entries related to your inputted LGG (Fig. 18). Additionally, by clicking on "view", you can view Kaplan-Meier curves between high-risk and low-risk groups in training and test data. Especially, REMR provide the time-dependent ROC curves assess the predictive performance of risk score based on triplets for survival time or event occurrence over time (Fig. 19).
- Similar to the queries in "TIPLETS" interface, you can query matching results by entering combinations of site, miRNA, gene, and cancer type according to your needs.
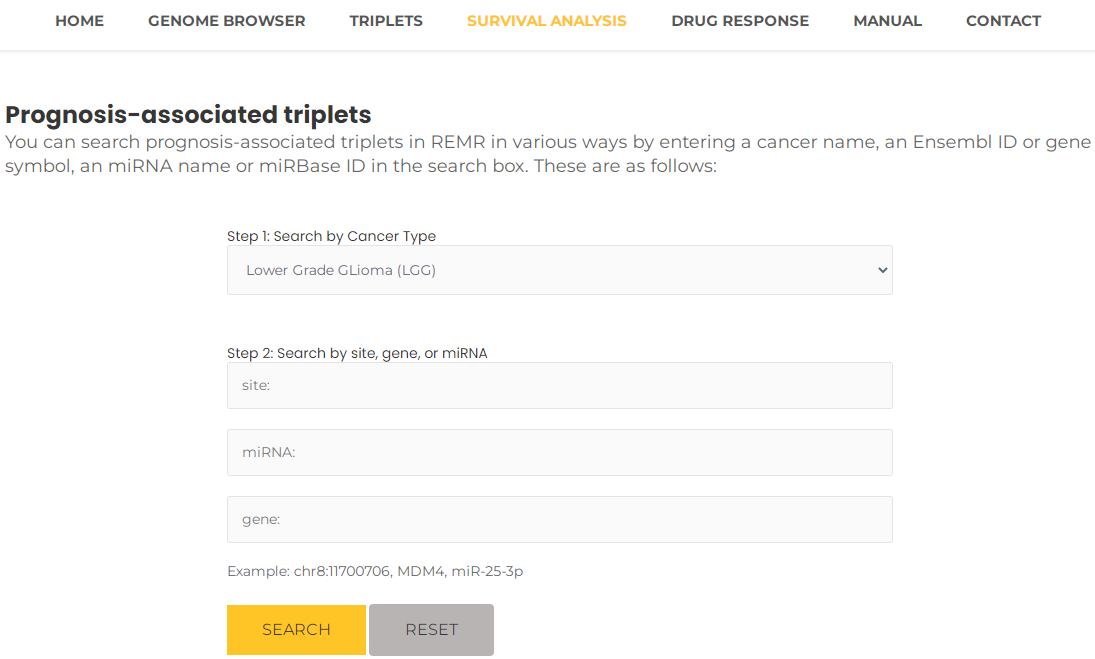
Fig.17 Search prognosis-associated triplets by "LGG".
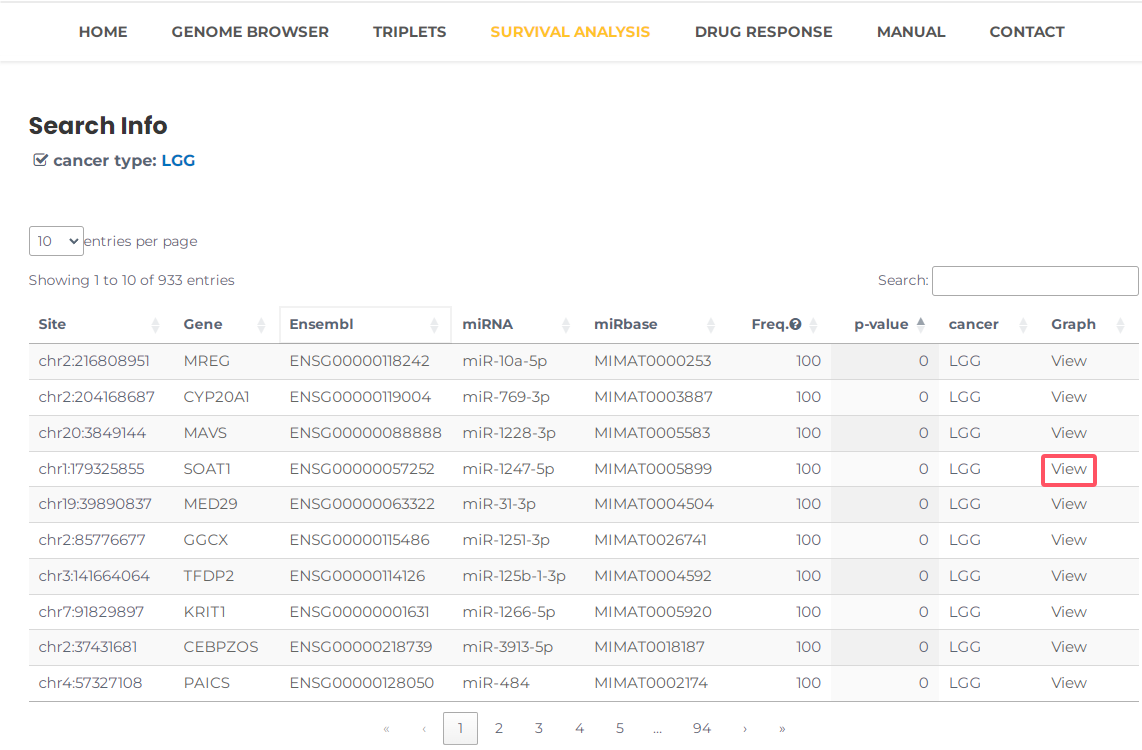
Fig.18 The result of "LGG".
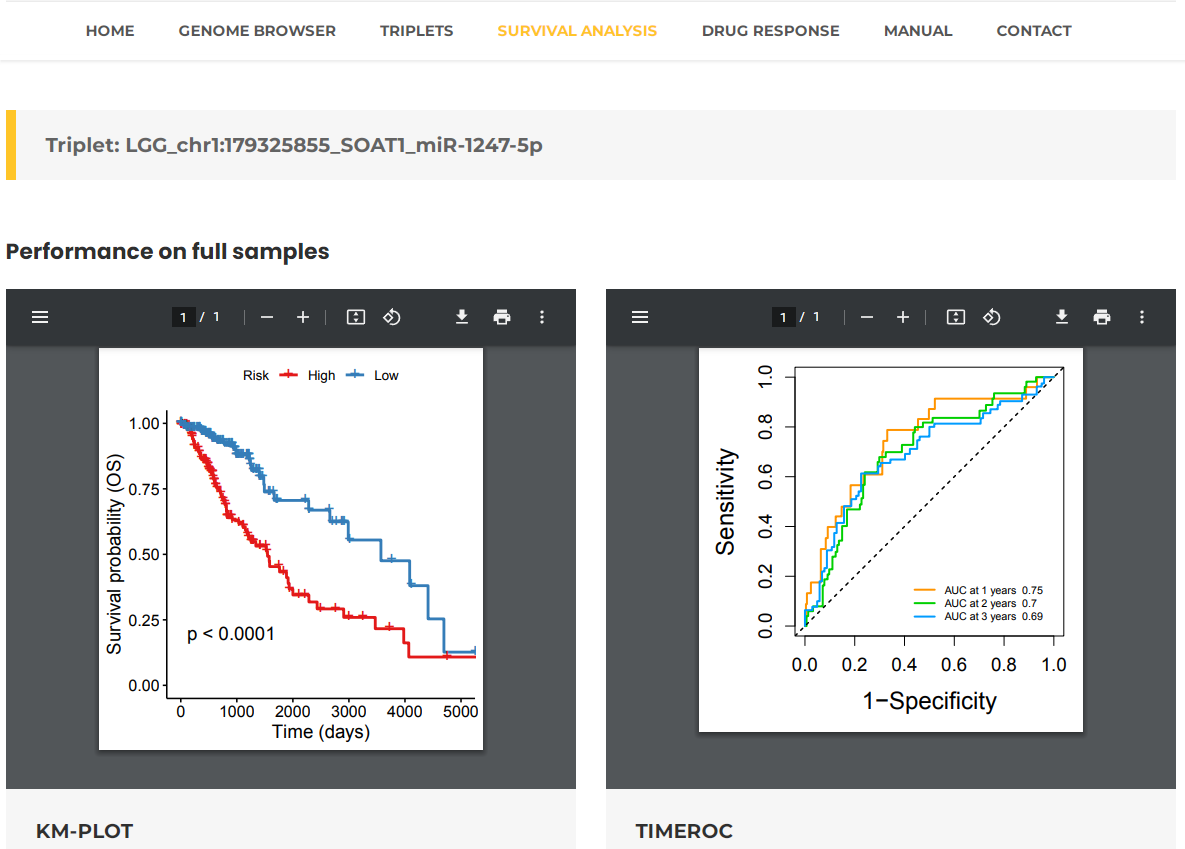
Fig.19 The KM-plot and timeROC of "chr1:179325855_SOAT1_miR-1247-5p" in "LGG".
In REMR database, you can review all identified drug response-associated triplets in a particular condition (one drug used in one cancer type).
- Taking the condition, Bladder Urothelial Carcinoma, BLCA_Gemcitabine, as an example. Selecting Bladder Urothelial Carcinoma, BLCA_Gemcitabine and then press the "Search" button (Fig. 20). The results will list all potential entries related to the triplet models that can effectively predict drug response for BLCA_Gemcitabine (Fig. 21).
Fig.20 Search drug response-associated triplets by condition.
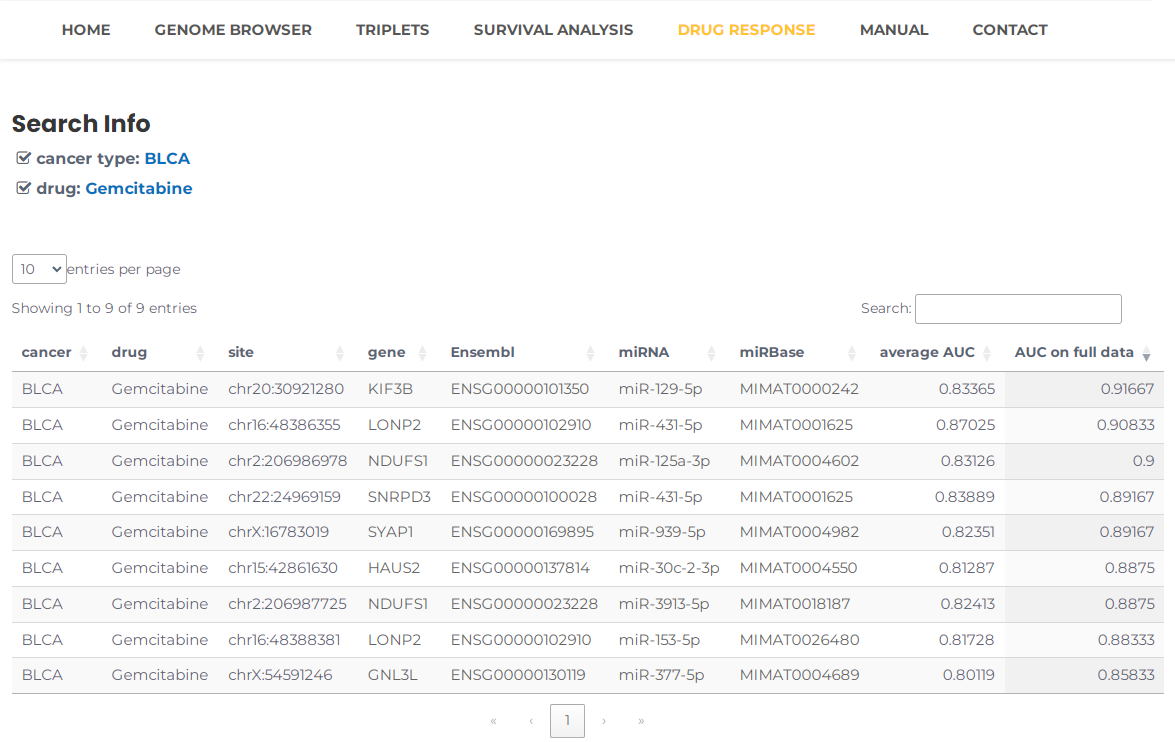
Fig.21 The result of "BLCA_Gemcitabine".
